The Metastatic streaming engine was developed to identify inpatient encounters with high risk of metastatic disease (cancer that spreads from where it started to a distant part of the body) and optimize the capture rate. It was deployed at The Mount Sinai Hospital on April 2022.
Clinical Documentation Integrity (CDI)
-
Core of every patient encounter is clinical documentation accurately reflecting the patient’s disease burden and scope of services provided.
-
Clinical documentation must be :
-
Clear
-
Consistent
-
Complete
-
Precise
-
Reliable
-
Timely
-
Legible
-
-
CDI facilitates the accurately translation of patient’s clinical status into coded data resulting in quality reporting, physician report cards, reimbursement, public health data, disease tracking and trending, and medical research.
-
The convergence of clinical care , documentation , and coding process is crucial for appropriate reimbursement, accurate quality scores, and informed decision-making to support high-quality patient care.
Challenges
-
High volume of clinical notes and the cost of processing
-
Manual chart review of cancer patients to identify new metastatic disease is inefficient due to
-
time required
-
limited number of patients assessed
-
difficulty identifying these patients prior to treatment
-
-
Information to quickly and accurately identify patients with metastatic disease is typically available only in clinical text documents (particularly radiology reports)
-
Complexity of language expression and inconclusive text to express uncertain or negative condition makes the NLP task very challenging 1
-
Building an exhaustive list of terms and rules to model language and extract domain concepts is extremely time consuming
-
High class-imbalance => low productivity
-
Clinical documentation improvement opportunity based on benchmarking reports
Current Approach and Solution
-
Search Algorithms
-
term/string matching and document indexing (
-
-
-
)
-
“metastatic”, “metastasis”, “metastases” and “carcinomatosis”
-
-
DNNs for medical NLP (Language models: embeddings)
-
BioBERT (
-
-
)
-
BiLSTM-CRF (
-
-
)
-
Relation Extraction (REX)2
-
-
Lexicon Mediated Entropy Reduction (LEXIMER) system
-
Medical Language Extraction and Encoding Sys- tem (MEDLEE)
-
It uses a controlled vocabulary and grammatical rules to translate text to a structured database format
-
Low generalizability 3,4
-
-
Radiology Analysis tool (RADA) 5
-
Mayo Clinic’s Clinical Text Analysis and Knowledge Extraction System (cTAKES)
-
a dictionary-based named-entity recognizer to highlight the Unified Medical Language System (UMLS) Metathesaurus terms in text, in addition to other NLP functionalities, such as tok- enizing, part of speech tagging, and parsing 6
-
-
Health Information Text Extraction (HITEx) from Brigham and Women’s Hospital and Harvard Medical School
-
It Creates tag for principal diagnoses 7
-
-
Named Entity Recognition (NER)2
-
Methods
-
dictionary-based method
-
conditional Markov model (CMM) 13
-
Sequence classifier
-
probabilities in CMMs are normalized locally for each state in the sequence
-
-
conditional random field model (CRF)
-
Sequence classifier
-
Probabilities in CRFs are normalized globally for a sequence
-
-
Information model 8,9
-
Structure
-
anatomy: “Right upper lobe”
-
anatomy modifier: “Anterior”
-
observation: “Mass”
-
observation modifier: “Calcified”,“1 cm”
-
uncertainty:“Probably is present”
-
-
This information model has a hierarchical structure
-
The annotation tool => eHOST:
-
-
-
eHOST: The Extensible Human Oracle Suite of Tools an open source annotation tool
-
Stanford Part of Speech Tagger
-
RadLex: RadLex lexicon is organized in a hierarchal structure and available in Web Ontology Language (OWL) format. 10
-
cTAKES dictionary-based named-entity recognition methodology in this work 12 (http://www.ohnlp.org/)
-
Using CMM and CRF train- ing infrastructure in Stanford Named-Entity Recognizer toolkit 11
Optimization opportunity and Goal
-
Opportunity: Identify inpatient encounters with high risk of metastatic disease and optimize the capture rate
-
Goal: Develop a ML based CDI tool to flag the inpatient encounters with high risk of metastatic disease at Discharge day and send the notification to the CDI specialist
Expected Impacts
-
Improve coding accuracy
-
Improve reimbursement opportunities
-
Improve comorbidity Score => Improve Elixhauser Comorbidity Index
-
Improve PSIs monitoring
Proposed Solution
-
This tool automatically screens patient’s clinical notes (Care Notes and Progress Notes) and reports (Radiology and Pathology) at discharge time for rapid identification of patients with metastatic disease
-
The machine learning information extraction approach provides an effective automatic method to annotate and extract clinically significant information from a large collection of free text and use a ML classifier to identify the patients with high risk of new metastasis
High Level Operationalization Workflow
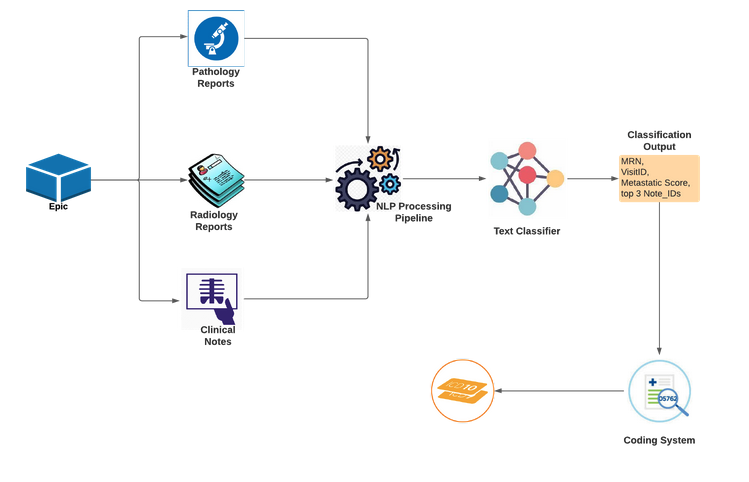
Batch Computational Flow
-
we use discriminative sequence classifiers for named-entity recognition to extract and organize clinically significant terms and phrases consistent with the information model.

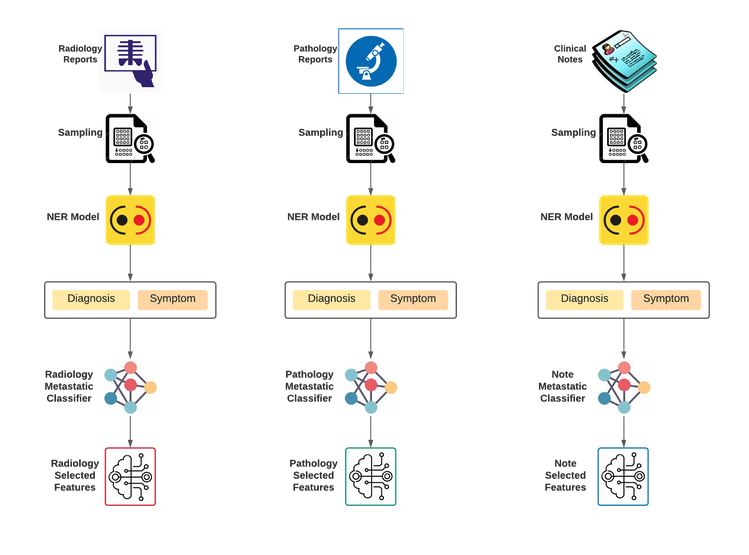
Feature Engineering Flow
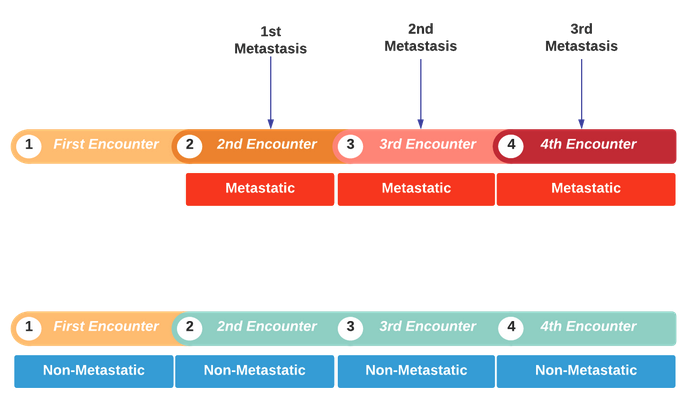
Labeling Logic
Proposed Key Performance Indicators (KPIs)
-
Chart review rate ==> will be captured by redcap response
-
Query rate
-
Provider response rate ==> will be captured by redcap response
-
Provider agreement rate ==> will be captured by redcap response
-
Unable to determine rate
Active Pilot Workflow
there are two types of metastatic patients:
-
documented and captured by 3M software
-
undocumented and will be captured by the CDI team review ==> only this category will be sent into the recap for being scanned by the NLP application

References
-
Chirag M Lakhani1 2, Arjun K Manrai1, 3, Jian Yang4, 5, Peter M Visscher#4, 5,*, and Chirag J Patel#1, 1Department BTT. 乳鼠心肌提取 HHS Public Access. Physiol Behav 2019;176:139–48. https://doi.org/10.1177/1535370213508172.Automated.
-
Hahn U, Oleynik M. Medical Information Extraction in the Age of Deep Learning. Yearb Med Inform 2020;29:208–20.
-
Hripcsak George, Kuperman Gilad J, Friedman Carol. Extracting findings from narrative reports: software transferability and sources of physician disagree- ment. Methods Inf Med 1998;37(1):1–7.
-
Elkins Jacob S, Friedman Carol, Boden-Albala Bernadette, Sacco Ralph L, Hripc- sak George. Coding neuroradiology reports for the Northern Manhattan Stroke Study: a comparison of natural language processing and manual review. Com- put Biomed Res 2000;33(1):1–10.
-
Johnson David B, Taira Ricky K, Cardenas Alfonso F, Aberle Denise R. Extract- ing information from free text radiology reports. Int J Digit Libr 1997;1(3): 297–308
-
Savova Guergana K, Masanz James J, Ogren Philip V, Zheng Jiaping, Sohn Sungh- wan, Kipper-Schuler Karin C, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applica- tions. J Am Med Inf Assoc 2010;17(5):507–13.
-
Goryachev Sergey, Sordo Margarita, Zeng Qing T. A suite of natural language processing tools developed for the I2B2 project. In: Bates David W, editor. Proceedings of the AMIA symposium, vol. 2. Washington DC: American Medical Informatics Association; 2006. p. 931
-
Langlotz Curtis P, Lee Meininger. Enhancing the expressiveness and usability of structured image reporting systems. In: Marc Overhage J, editor. Proceedings of the AMIA symposium. Los Angeles, CA: American Medical Informatics Asso- ciation; 2000. p. 467
-
Hassanpour S, Langlotz CP. Information extraction from multi-institutional radiology reports. Artif Intell Med 2016;66:29–39.
-
Langlotz Curtis P. RadLex: a new method for indexing online educational mate- rials. Radiographics 2006;26(6):1595–7
-
Finkel Jenny Rose, Grenager Trond, Manning Christopher. Incorporating non- local information into information extraction systems by Gibbs sampling. In: Darwish Kareem, Diab Mona, Habash Nizar, editors. Proceedings of the 43rd annual meeting on association for computational linguistics. Ann Arbor, MI: Association for Computational Linguistics; 2005. p. 363–70
-
Savova Guergana K, Masanz James J, Ogren Philip V, Zheng Jiaping, Sohn Sungh- wan, Kipper-Schuler Karin C, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applica- tions. J Am Med Inf Assoc 2010;17(5):507–13
-
Ratnaparkhi Adwait. A maximum entropy model for part-of-speech tagging. In: Brill Eric, Church Kenneth, editors. Proceedings of the conference on empirical methods in natural language processing, vol. 1. Philadelphia, PA: Association for Computational Linguistics; 1996. p. 133–42